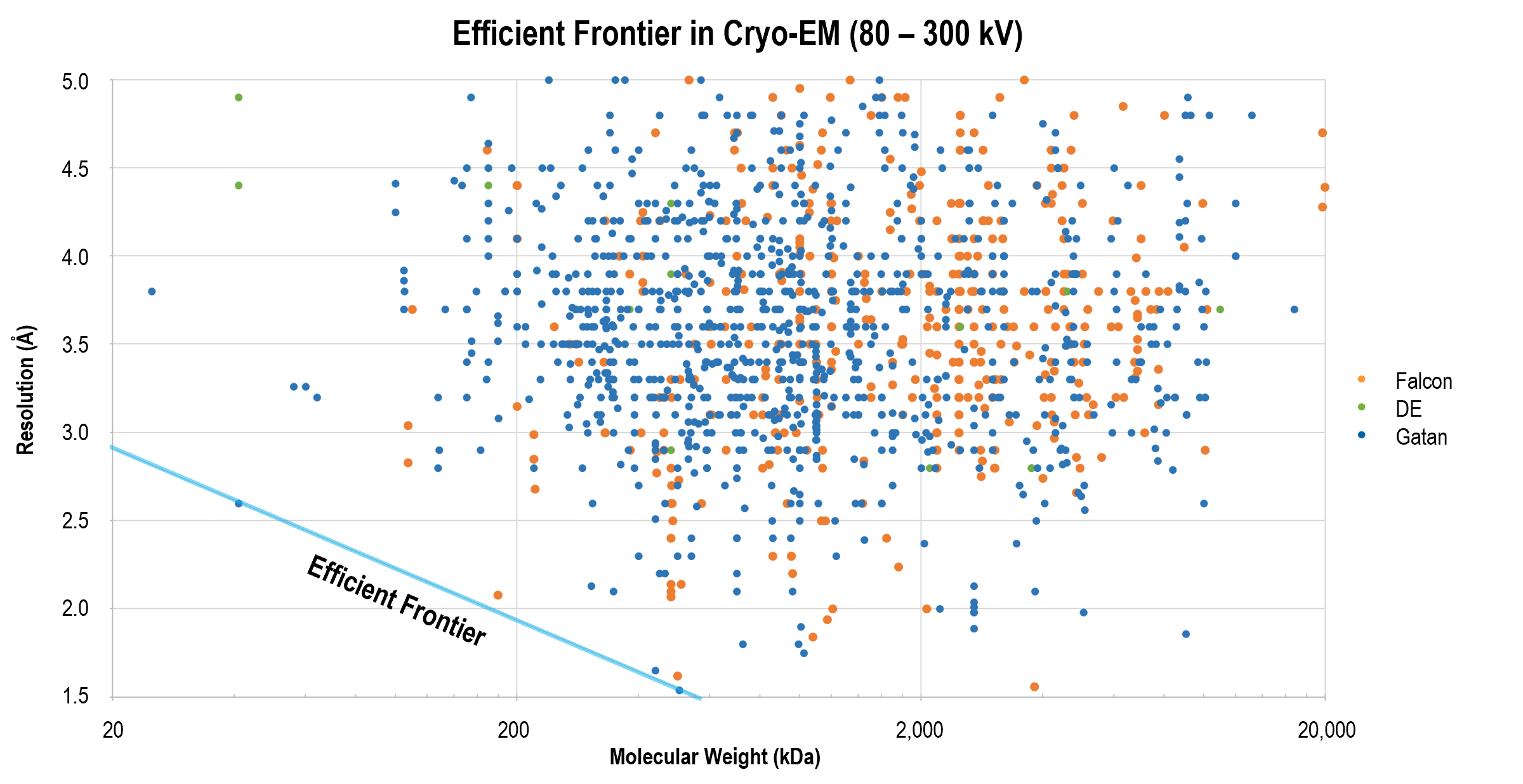
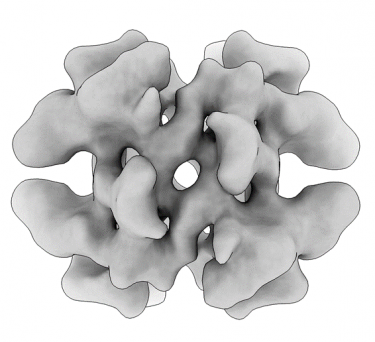
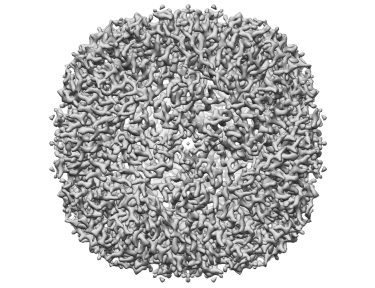
Obtaining high-resolution cryo-EM structures using a common LaB6, 120-keV electron microscope equipped with a sub 200-keV optimised direct electron detector
Venugopal, H.; Mobbs, J.; Taveneau, C.; Fox, D. R.; Vuckovic, Z.; Knott, G.; Grinter, R.; Thal, D.; Mick, S.; Czarnik, C.; Ramm, G.
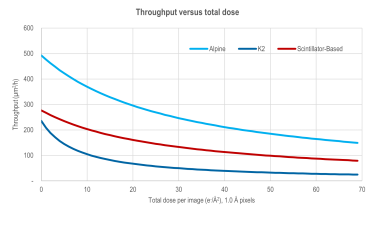
High-resolution single particle imaging at 100-200 keV with the Gatan Alpine direct electron detector
Chan, L. M.; Courteau, B. J.; Maker, A.; Wu, M.; Basanta, B.; Mehmood, H.; Bulkley, D.; Joyce, D.; Lee, B. C.; Mick, S.; Gulati, S.; Lander, G. C.; Verba, K. A.
Helical ultrastructure of the metalloprotease meprin α in complex with a small molecule inhibitor
Bayly-Jones, C.; Lupton, C. J.; Fritz, C.; Venugopal, H.; Ramsbeck, D.; Wermann, M.; Jäger,C.; Marco, A. D.; Schilling, S.; Schlenzig, D.; Whisstock, J. C.
Abundant Aβ fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer’s disease brains
Stern, A. M.; Yang, Y.; Meunier, A. L.; Liu, W.; Cai, Y.; Ericsson, M.; Liu, L.; Goedert, M.; Scheres, S. H. W.; Selkoe, D.J
Cryo-EM structure of the human NKCC1 transporter reveals mechanisms of ion coupling and specificity
Neumann, C.; Rosenbæk, L. L.; Flygaard, R. K.; Habeck, M.; Karlsen, J. L.; Wang, Y.; Larsen, K. L.; Gad, H. H.; Hartmann, R.; Lyons, J. A; Fenton, R. A.; Nissen, P.
Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex
Naschberger, A.; Mosebach, L.; Tobiasson, V.; Kuhlgert, S.; Scholz, M.; Perez-Boerema, A.; Ho, T. T. H.; Vidal-Meireles, A.; Takahashi, Y.; Hippler, M.; Amunts, A.
IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern
Chang, M. R.; Tomasovic, L.; Kuzmina, N. A.; Ronk, A. J.; Byrne, P. O.; Johnson, R.; Storm, N.; Olmedillas, E.; Hou, Y. J.; Schäfer, A.; Leist, S. R.; Tse, L. T.; Ke, H.; Coherd, C.; Nguyen, K.; Kamkaew, M.; Honko, A.; Zhu, Q.; Alter, G.; Saphire, E. O.; McLellan, J. S.; Griffiths, A.; Baric, R. S.; Bukreyev. A.; Marasco. W. A.
Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Judge, R. A.; Sridar, J.; Tunyasunvunakool, K.; Jain, R.; Wang, J. C. K.; Ouch, C.; Xu, J.; Mafi, A.; Nile, A. H.; Remarcik, C.; Smith, C. L.; Ghosh, C.; Xu, C.; Stoll, V.; Jumper, J.; Singh, A. H.; Eaton, D.; Hao, Q.
Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase
Miyakawa, T.; Yang, J.; Kawasaki, M.; Adachi, N.; Fujii, A.; Miyauchi, Y.; Muramatsu, T.; Moriya, T.; Senda, T.; Tanokura, M.
Structural insights into dsRNA processing by Drosophila Dicer-2–Loqs-PD
Su, S.; Wang, J.; Deng, T.; Yuan, X.; He, J.; Liu, N.; Li, X.; Huang, Y.; Wang, H. -W.; Ma, J.
Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike
Stalls, V.; Lindenberger, J.; Gobeil, S. M. -C.; Henderson, R.; Parks, R.; Barr, M.; Deyton, M.; Martin, M.; Janowska, K.; Huang, X.; May, A.;l Speakman, M.; Beaudoin, E.; Kraft, B.; Lu, X.;
Edwards, R. J.; Eaton, A.; Montefiori, D. C.; Williams, W.; Saunders, K. O.; Wiehe, K.; Haynes, B. F.; Acharya, P.
Structure of the type V-C CRISPR-Cas effector enzyme
Kurihara, N.; Nakagawa, R.; Hirano, H.; Okazaki, S.; Tomita, A.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Scott, D. A.; Nishimasu, H.; Nureki, O.
The giant Mimivirus 1.2 Mb genome is elegantly organized into a 30 nm helical protein shield
Villalta, A.; Schmitt, A.; Estrozi, L. F.; Quemin, E. R. J.; Alempic, J. -M.; Lartigue, A.; Pražák, V.; Belmudes, L.; Vasishtan, D.; Colmant, A. M. G.; Honoré, F. A.; Couté, Y.; Grünewald, K.; Abergel, C.
Structural and functional impact by SARS-CoV-2 Omicron spike mutations
Zhang, J.; Cai, Y.; Lavine, C. L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M. L.; Rits-Volloch, S.; Wang, S.; Sliz, P.; Wesemann, D. R.; Yang, W.; Seaman, M. S.; Lu, J.; Xiao, T.; Chen, B.
Capturing the swelling of solid-electrolyte interphase in lithium metal batteries
Zhang, Z.; Li, Y.; Xu, R.; Zhou, W.; Li, Y.; Oyakhire, S. T.; Wu, Y.; Xu, J.; Wang, H.; Yu, Z.; Boyle, D. T.; Huang, W.; Ye, Y.; Chen, H.; Wan, J.; Bao, Z.; Chiu, W.; Cui, Y.
Nuclear pores dilate and constrict in cellulo
Zimmerli, C. E.; Allegretti, M.; Rantos, V.; Goetz, S. K.; Obarska-Kosinska, A.; Zagoriy, I.; Halavatyi, A.; Hummer, G.; Mahamid, J.; Kosinski, J.; Beck, M.
Structural basis and mechanism of activation of two different families of G proteins by the same GPCR
Nature Structural & Molecular Biology
Alegre, K. O.; Paknejad, N.; Su, M.; Lou, J. -S.; Huang, J.; Jordan, K. D.; Eng, E. T.; Meyerson, J. R.; Hite, R. K.; Huang, X. -Y.
Structurally mapping antigenic epitopes of adeno-associated virus 9: Development of antibody escape variants
Emmanuel, S. N.; Smith, J. K.; Hsi, J.; Tseng, Y. -S.; Kaplan, M.; Mietzsch, M.; Chipman, P.; Asokan, A.; Robert McKenna, R.; Agbandje-McKenna, M.
Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2
Peng, Q.; Peng, R.; Yuan, B.; Zhao, J.; Wang, M.; Wang, X.; Wang, Q.; Sun, Y.; Fan, Z.; Qi, J.; Gao, G. F.; Shi, Y.
Native-like SARS-CoV-2 spike glycoprotein expressed by ChAdOx1 nCoV-19/AZD1222 vaccine
Watanabe, Y.; Mendonça, L.; Allen, E. R.; Howe, A.; Lee, M.; Allen, J. D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; Ulaszewska, M.; Belij-Rammerstorfer, S.; Morris, S.; Krebs, A. -S.; Dejnirattisai, W.; Mongkolsapaya, J.; Supasa, P.; Screaton, G. R.; Green, C. M.; Lambe, T.; Zhang, P.; Gilbert, S. C.; Crispin, M.
Mechanism of SARS-CoV-2 polymerase stalling by remdesivir
Kokic, G.; Hillen, H. S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P.
Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Koenig, P. -D.; Das, H.; Liu H.; Kümmerer, B. M.; Gohr, F. N.; Jenster, L. -M.; Schiffelers, L. D. J.; Tesfamariam, Y. M.; Uchima, M.; Wuerth, J. D.; Gatterdam, K.; Ruetalo, N.; Christensen, M. H.; Fandrey, C. I.; Normann, S.; Tödtmann, J.; M. P.; Pritzl, S.; Hanke, L.; Boos, J.; Yuan, M.; Zhu, X.; Schmid-Burgk, J. L.; Kato, H.; Schindler, M.; Wilson, I. A.; Geyer, M.; Ludwig, K. U.; Hällberg, M.; Wu, N. C.; Schmidt, F. I.
Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections
Zhang, C.; Wang, Y.; Zhu, Y.; Liu, C.; Gu, C.; Xu, S.; Wang, Y.; Zhou, Y.; Wang, Y.; Han, W.; Hong, X.; Yang, Y.; Zhang, X.; Wang, T.; Xu, C.; Hong, Q.; Wang, S.; Zhao, Q.; Qiao, W.; Zang, J.; Kong, L.; Wang, F.; Wang, H.; Qu, D.; Lavillette, D.; Tang, H.; Deng, Q.; Xie, Y.; Cong, Y.; Huang, Z.
Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis
Yan, L.; Ge, J.; Zheng, L.; Zhang, Y.; Gao, Y.; Wang, T.; Huang, Y.; Yang, Y.; Gao, S.; Li, M.; Liu, Z.; Wang, H.; Li, Y.; Chen, Y.; Guddat, L. W.; Wang, Q.; Rao, Z.; Lou, Z.
Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM
Xu, C.; Wang, Y.; Liu, C.; Zhang, C.; Han, W.; Hong, X.; Wang, Y.; Hong, Q.; Wang, S.; Zhao, Q.; Wang, Y.; Yang, Y.; Chen, K.; Zheng, W.; Kong, L.; Wang, F.; Zuo, Q.; Huang, Z.; Cong, Y.
Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA
Yuan, S.; Peng, L.; Park, J. J.; Hu, Y.; Devarkar, S. C.; Dong, M. B.; Shen, Q.; Wu, S.; Chen, S.; Lomakin, I. B.; Xiong, Y.
Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains
Zhou, T.; Tsybovsky, Y.; Gorman, J.; Rapp, M.; Cerutti, G.; Chuang, G. -Y.; Katsamba, P. S.; Sampson, J. M.; Schön, A.; Bimela, J.; Boyington, J. C.; Nazzari, A.; Olia, A. S.; Shi, W.; Sastry, M.; Stephens, T.; Stuckey, J.; Teng, I. -T.; Kwong, P. D
De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2
Linsky, T. W.; Vergara, R.; Codina, N.; Nelson, J. W.; Walker, M. J.; Su, W.; Barnes, C. O.; Hsiang, T. -Y.; Esser-Nobis, K.; Yu, K.; Reneer, B.; Hou, Y. J.; Priya, T.; Mitsumoto, M.; Pong, A.; Lau, Y.; Mason, M. L.; Chen, J.; Chen, A.; Berrocal, T.; Peng, H.; Clairmont, N. S.; Castellanos, J.; Lin, Y. -R..; Josephson-Day, A.; Baric, R. S.; Fuller, D. H.; Walkey, C. D.; Ross, T. M.; Swanson, R.; Bjorkman, P. J.; Gale Jr., M.; Blancas-Mejia, L. M.; Yen, H. -L.; Silva, D. -A.
Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection
Yao, H.; Sun, Y.; Deng, Y. -Q.; Wang, N.; Tan, Y.; Zhang, N. -N.; Li, X. -F.; Kong, C.; Xu, Y. -P.; Chen, Q.; Cao, T. -S.; Zhao, H.; Yan, X.; Cao, L.; Lv, Z.; Zhu, D.; Feng, R.; Wu, N.; Zhang, W.; Hu, Y.; Chen, K.; Zhang, R. -R.; Lv, Q.; Sun, S.; Zhou, Y.; Yan, R.; Yang, G.; Sun, X.; Liu, C.; Lu, X.; Cheng, L.; Qiu, H.; Huang, X. -Y.; Weng, T.; Shi, D.; Jiang, W.; Shao, J.; Wang, L.; Zhang, J.; Jiang, T.; Lang, G.; Qin, C. -F.; Li, L.; Wang, X.
Structure-based development of human antibody cocktails against SARS-CoV-2
Wang, N.; Sun, Y.; Feng, R.; Wang, Y.; Guo, Y.; Zhang, L.; Deng, Y. -Q.; Wang, L.; Cui, Z.; Cao, L.; Zhang, Y. -J.; Li, W.; Zhu, F. -C.; Qin, C. -F.; Wang, X.
Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate
Bangaru, S.; Ozorowski, G.; Turner, H. L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J. L.; Diedrich, J. K.; Tian, J. -H.; Portnoff, A. D.; Patel, N.; Massare, M. J.; Yates III, J. R.; Nemazee, D.; Paulson, J. C.; Glenn, G.; Smith, G.; Ward, A. B.
Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms
Tortorici, M. A.; Beltramello, M.; Lempp, F. A.; Pinto, D.; Dang, H. V.; Rosen, L. E.; McCallum, M.; Bowen, J.; Minola, A.; Jaconi, S.; Zatta, F.; De Marco, A.; Guarino, B.; Bianchi, S.; Lauron, E. J.; Tucker, H.; Zhou, J.; Peter, A.; Havenar-Daughton, C.; Wojcechowskyj, J. A.; Case, J. B.; Chen, R. E.; Kaiser, H.; Montiel-Ruiz, M.; Meury, M.; Czudnochowski, N.; Spreafico, R.; Dillen, J.; Ng, C.; Sprugasci, N.; Culap, K.; Benigni, F.; Abdelnabi, R.; Foo, S. -Y. C.; Schmid, M. A.; Cameroni, E.; Riva, A.; Gabrieli, A.; Galli, M.; Pizzuto. M. S.; Neyts, J.; Diamond, M. S.; Virgin, H. W.; Snell, G.; Corti, D.; Fink, K.; Veesler, D.;
SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography
Klein, S.; Cortese, M.; Winter, S. L.; Wachsmuth-Melm, M.; Neufeldt, C. J.; Cerikan, B.; Stanifer, M. L.; Boulant, S.; Bartenschlager, R.; Chlanda , P.
Architecture of a SARS-CoV-2 mini replication and transcription complex
Yan, L.; Zhang, Y.; Ge, J.; Zheng, L.; Gao, Y.; Wang, T.; Jia, Z.; Wang, H.; Huang, Y.; Li, M.; Wang, Q.; Ra, Z.; Lou, Z.
Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology
Piccoli, L.; Park, Y. -J.; Tortorici, M. A.; Czudnochowski, N.; Walls, A. C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L. E.; Bowen, J. E.; Acton, O. J.; Jaconi, S.; Guarino, B.; Minola, A.; Zatta, F.; Sprugasci, N.; Bassi, J.; Peter, A.; De Marco, A.; Nix, J. C.; Mele, F.; Jovic, S.; Rodriguez, B. F.; Gupta, S. V.; Jin, F.; Piumatti, G.; Presti, G. L.; Pellanda, A. F.; Biggiogero, M.; Tarkowski, M.; Pizzuto, M. S.; Cameroni, E.; Havenar-Daughton, C.; Smithey, M.; Hong, D.; Lepori, V.; Albanese, E.; Ceschi, A.; Bernasconi, E.; Elzi, L.; Ferrari, P.; Garzoni, C.; Riva, A.; Snell, G.; Sallusto, F.; Fink, K.; Virgin, H. W.; Lanzavecchia, A.; Corti, D.; Veesler, D.
Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy
Du, S.; Cao, Y.; Zhu, Q.; Yu, P.; Qi, F.; Wang, G.; Du, X.; Bao, L.; Deng, W.; Zhu, H.; Liu, J.; Nie, J.; Zheng, Y.; Liang, H.; Liu, R.; Gong, S.; Xu, H.; Yisimayi, A.; Qin, C.
Engineered trimeric ACE2 binds viral spike protein and locks it in “Three-up” conformation to potently inhibit SARS-CoV-2 infection
Guo, L.; Bi, W.; Wang, X.; Xu, W.; Yan, R.; Zhang, Y.; Zhao, K.; Li, Y.; Zhang, M.; Cai, X.; Jiang, S.; Xie, Y.; Zhou, Q.; Lu, L.; Dang, B.
Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein
Toelzer, C.; Gupta, K.; Yadav, S. K. N.: Borucu, U.; Davidson, A. D.; Williamson, M. K.; Shoemark, D. K.; Garzoni, F.; Staufer, O.; Milligan, R.; Capin, J.; Mulholland, A. J.; Spatz, J.; Fitzgerald, D.; Berger, I.; Schaffitzel, C.
An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive spike
Schoof, M.; Faust, B.; Saunders, R. A.; Sangwan, S.; Rezelj, V.; Hoppe, N.; Boone, M.; Billesbølle, C. B.; Puchades, C.; Azumaya, C. M.; Kratochvil, H. T.; Zimanyi, M.; Deshpande, I.; Liang, J.; Dickinson, S.; Nguyen, H. C.; Chio, C. M.; Merz, G. E.; Thompson, M. C.; Diwanji, D.; Schaefer, K.; Anand, A. A.; Dobzinski, N.; Zha, B. S.; Simoneau, C. R.; Leon, K.; White, K. M.; Chio, U. S.; Gupta, M.; Jin, M.; Li, F.; Liu, Y.; Zhang, K.; Bulkley, D.; Sun, M.; Smith, A. M.; Rizo, A. N.; Moss, F.; Brilot, A. F.; Pourmal, S.; Trenker, R.; Pospiech, T.; Gupta, S.; Barsi-Rhyne, B.; Belyy, V.; Barile-Hill, A. W.; Nock, S.; Liu, Y.; Krogan, N. J.; Ralston, C. Y.; Swaney, D. L.; García-Sastre, A.; Ott, M.; Vignuzzi, M.; QCRG Structural Biology Consortium; Walter, P.; Manglik, A.
Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2
Custódio, T. F.; Das, H.; Sheward, D. J.; Hanke, L.; Pazicky, S.; Pieprzyk, J.; Sorgenfrei, M.; Schroer, M. A.; Gruzinov, A. Y.; Jeffries, C. M.; Graewert, M. A.; Svergun, D. I.; Dobrev, N.; Remans, K.; Seeger, M. A.; McInerney, G. M.; Murrell, B.; Hällberg, B. M.; Löw, C.
The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by cryo-EM and cryo-ET
Liu, C.; Mendonça, L.; Yang, Y.; Gao, Y.; Shen, C.; Liu, J.; Ni, T.; Ju, B.; Liu, C.; Tang, X.; Wei, J.; Ma, X.; Zhu, Y.; Liu, W.; Xu, S.; Liu, Y.; Yuan, J.; Wu, J.; Liu, Z.; Zhang, Z.; Liu, L.; Wang, P.; Zhang, P.
Structure-based design with tag-based purification and in-process biotinylation enable streamlined development of SARS-CoV-2 spike molecular probes
Zhou, T.; Teng, I. -T.; Olia, A. S.; Cerutti, G.; Gorman, J.; Nazzari, A.; Shi, W.; Tsybovsky, Y.; Wang, L.; Wang, S.; Zhang, B.; Zhang, Y.; Katsamba, P. S.; Petrova, Y.; Banach, B. B.; Fahad, A. S.; Liu, L.; Lopez Acevedo, S. N.; Madan, B.; de Souza, M. O.; Pan, X.; Wang, P.; Wolfe, J. R.; Yin, M.; Ho, D. D.; Phung, E.; DiPiazza, A.; Chang, L. A.; Abiona, O. M.; Corbett, K. S.; DeKosky, B. J.; Graham, B. S.; Mascola, J. R.; Misasi, J.; Ruckwardt, T.; Sullivan, N. J.; Shapiro, L.; Kwong, P. D.
De novo design of picomolar SARS-CoV-2 miniprotein inhibitors
Cao, L.; Goreshnik, I.; Coventry, B.; Case, J. B.; Miller, L.; Kozodoy, L.; Chen, R. R.; Carter, L.; Walls, A. C.; Park, Y. -J.; Strauch, E. -M.; Stewart, L.; Diamond, M. S.; Veesler, D.; Baker, D.
SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies
Barnes, C. O.; Jette, C. A.; Abernathy, M. E.; Dam, K. -M., A.; Esswein, S. R.; Gristick, H. B.; Malyutin, A. G.; Sharaf, N. G.; Huey-Tubman, K. E.; Lee, Y. E.; Robbiani, D. F.; Nussenzweig, M. C.; West Jr., A. P.; Bjorkman, P. J.
Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2
Wu, L.; Chen, Q.; Liu, K.; Wang, J.; Han, P.; Zhang, Y.; Hu, Y.; Meng, Y.; Pan, X.; Qiao, C.; Tian, S.; Du, P.; Song, H.; Shi, W.; Qi, J.; Wang, H. -W.; Yan, J.; Gao, G. F.; Wang, Q.
Structure-based design of prefusion-stabilized SARS-CoV-2 spikes
Hseih, C. -L.; Goldsmith, J. A.; Schaub, J. M.; Divenere, A. M.; Kuo, H. -C.; Javanmardi, K.; Le, K. C.; Wrapp, D.; Lee, A. G.; Liu, Y.; Chou, C. -W.; Byrne, P. O.; Hjorth, C. K.; Johnson, N. V.; Ludes-Meyers, J.; Nguyen, A. W.; Park, J.; Wang, N.; Amengor, D.; Lavinder, J. J.; Ippolito, G. C.; Maynard, J. A.; Finkelstein, I. J.; McLellan, J. S.
Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody
Lv, Z.; Deng, Y. -Q.; Ye, Q.; Cao, L.; Sun, C. -Y.; Fan, C.; Huang, W.; Sun, S.; Sun, Y.; Zhu, L.; Chen, Q.; Wang, N.; Nie, J.; Cui, Z.; Zhu, D.; Shaw, N.; Li, X. -F.; Li, Q.; Xie, L.; Wang, Y.; Rao, Z.; Qin, C. -F.; Wang, X.
Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex
Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P. M. M.; Olinares, P. D. B.; Maruthi, K.; Eng, E. T.; Vatandaslar, H.; Chait, B. T.; Kapoor, T. M.; Darst, S. A.; Campbell, E. A.
SARS-CoV-2 Nsp1 binds the ribosomal mRMA channel to inhibit translation
Nature Structural & Molecular Biology
Schubert, K.; Karousis, E. D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L. -A.; Leibundgut, M.; Thiel, V.; Mühlemann, O.; Ban, N.
Neutralization of SARS-CoV-2 by destruction of the prefusion spike
Huo, J.; Zhao, Y.; Ren, J.; Zhou, D.; Duyvesteyn, H. M. E.; Ginn, H. M.; Carrique, L.; Malinauskas, T.; Ruza, R. R.; Shah, P. N. M.; Tan, T. K.; Rijal, P.; Coombes, N.; Bewley, K. R.; Tree, J. A.; Radecke, J.; Paterson, N. G.; Supasa, P.; Mongkolsapaya, J.; Screaton, G. R.; Carroll, M.; Townsend, A.; Fry, E. E.; Owens, R. J.; Stuart, D. I.
An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction
Hanke, L.; Perez, L. V.; Sheward, D. J.; Das, H.; Schulte, T.; Moliner-Morro, A.; Corcoran, M.; Achour, A.; Hedestam, G. B.; Hällberg, B. M.; Murrell, B.; McInerney, G. M.
Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2
Thoms, M.; Buschauer, R.; Ameismeier, M.; Loepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; Straub, J. H.; Sturzel, C. M.; Frohlich, T.; Berninghausen, O.; Becker, T.; Kirchhoff, F.; Sparrer, K. M.; Beckmann, R.
Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail
Hansen, J.; Baum, A.; Pascal, K. E.; Russo, V.; Giordano, S.; Wloga, E.; Fulton, B. O.; Yan, Y.; Koon, K.; Patel, K.; Chung, K. M.; ermann, A.; Ullman, E.; Cruz, J.; Rafique, A.; Huang, T.; Fairhurst, J.; Libertiny, C.; Malbec, M.; Lee, W. -Y.; Welsh, R.; Farr, G.; Pennington, S.; Deshpande, D.; Cheng, J.; Watty, A.; Bouffard, P.; Babb, R.; Levenkova, N.; Chen, C.; Zhang, B.; Hernandez, A. R.; Saotome, K.; Zhou, Y.; Franklin, M.; Sivapalasingam, S.; Lye, D. C.; Weston, S.; Lopgue, J.; Haupt, R.; Frieman, M.; Chen., G.; Olson, W.; Murphy, A. J.; Stahl, N.; Yancopoulos, G. D.; Kyratsous, C. A.
Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies
Barnes, C. O.; West Jr., A. P.; Huey-Tubman, K. E.; Hoffmann, M. A. G.; Sharaf, N. G.; Hoffman, P. R.; Koranda, N.; Gristick, H. B.; Gaebler, C.; Muecksch, F.; Lorenzi, J. C. C.; Finkin, S.; Hägglöf, T.; Hurley, A.; Millard, K. G.; Weisblum, Y.; Schmidt, F.; Hatziioannou, T.; Bieniasz, P. D.; Caskey, M.; Robbiani, D. F.; Nussenzweig, M. C.; Bjorkman. P. J.
A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2
Chi, X.; Yan, R.; Zhang, G.; Zhang, Z.; Hao, M.; Zhang, Z.; Fan, P.; Dong, X.; Yang, Y.; Chen, Z.; Guo, Y.; Zhang, J.; Li, Y.; Song, X.; Cheng, Y.; Xia, L.; Fu, L.; Hou, L.; Xu, J.; Yu, X.; Li, J.; Zhou, Q.; Chen, W.
Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient
Nature Structural & Molecular Biology
Zhou, D.; Duyvesteyn, H. M. E.; Chen, C. -P.; Huang, C. -G.; Chen, T. -H.; Shih, S. -R.; Lin, Y. -C.; Cheng, C. -Y.; Cheng, S. -H.; Huang, Y. -C.; Lin, T. -Y.; Ma, C.; Huo, J.; Carrique, L.; Malinauskas, T.; Ruza, R. R.; Shah, P. N. M.; Tan, T. K.; Rijal, P.; Donat, R. F.; Godwin, K.; Buttigieg, K. R.; Tree, J. A.; Radecke, J.; Paterson, N. G.; Supasa, P.; Mongkolsapaya, J.; Screaton, G. R.; Carroll, M. W.; Gilbert-Jaramillo, J.; Knight, M. L.; James, W.; Owens, R. J.; Naismith, J. H.; Townsend, A. R.; Fry, E. E.; Zhao, Y.; Ren, J.; Stuart, D. I.; Huang, K. -Y. A.
A thermostable, closed SARS-CoV-2 spike protein trimer
Nature Structural & Molecular Biology
Xiong, X.; Qu, K.; Ciazynska, K. A.; Hosmillo, M.; Carter, A. P.; Ebrahimi, S.; Ke, Z.; Scheres, S. H. W.; Bergamaschi, L.; Grice, G. L.; Zhang, Y.; The CITIID-NIHR COVID-19 BioResource Collaboration; Nathan, J. A.; Baker, S.; James, L. C.; Baxendale, H. E.; Goodfellow, I.; Doffinger, R.; Briggs, J. A. G.
Structural basis for RNA replication by the SARS-CoV-2 polymerase
Wang, Q.; Wu2, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; Fang, X.; Yang, X.; Huang, Y.; Gao, H.; Liu, F.; Ge, J.; Sun, Q.; Yang, X.; Xu, W.; Liu, Z.; Yang, H.; Lou, Z.; Jiang, B.; Guddat, L. W.; Gong, P.; Rao, Z.
Controlling the SARS-CoV-2 spike glycoprotein conformation
Nature Structural & Molecular Biology
Henderson, R.; Edwards, R. J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S. M. C.; Kopp, M.; Li, D.; Parks, R.; Hsu, A. L.; Borgnia, M. J.; Haynes, B. F.; Acharya, P.
Potently neutralizing and protective human antibodies against SARS-CoV-2
Zost, S. J.; Gilchuk, P.; Case, J. B.; Binshtein, E.; Chen, R. E.; Nkolola, J. P.; Schäfer, A.; Reidy, J. X.; Trivette, A.; Nargi, R. S.; Sutton, R. E.; Suryadevara, N.; Martinez, D. R.; Williamson, L. E.; Chen, E. C.; Jones, T.; Day, S.; Myers, L.; Hassan, A. O.; Kafai, N. M.; Winkler, E. S.; Fox, J. M.; Shrihari, S.; Mueller, B. K.; Meiler, J.; Chandrashekar, A.; Mercado, N. B.; Steinhardt, J. J.; Ren, K.; Loo, Y. -M.; Kallewaard, N. L.; McCune, B. T.; Keeler, S. P.; Holtzman, M. J.; Barouch, D. H.; Gralinski, L. E.; Baric, R. S.; Thackray, L. B.; Diamond, M. S.; Carnahan, R. H.; Crowe Jr, J. E.
Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells
Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; Chai, X.; He, R.; Li, X.; Lv, Q.; Zhu, H.; Deng, W.; Xu, Y.; Wang, Y.; Qiao, L.; Tan, Y.; Song, L.; Wang, G.; Du, X.; Gao, N.; Liu, J.; Xiao, J.; Su, X. -D.; Du, Z.; Feng, Y.; Qin, C.; Qin, C.; Jin, R.; Xie, X. S.
Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir
Yin, W.; Mao, C.; Luan, X.; Shen, D. -D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; Chang, S.; Xie, Y. -C.; Tian, G.; Jiang, H. -W.; Tao, S. -C.; Shen, J.; Jiang, Y.; Jiang, H.; X.u, Y.; Zhang, S.; Xu, H. E.
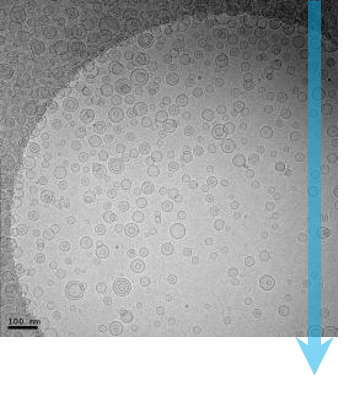
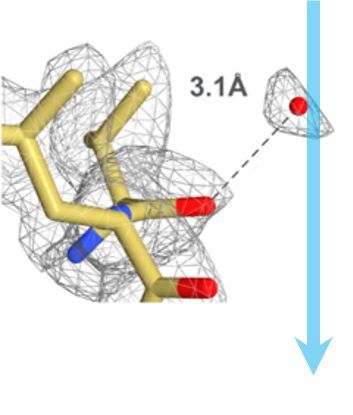
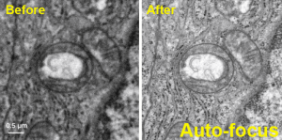
 Single-particle cryo-electron microscopy (cryo-EM), is an increasingly popular technique used by structural biologists to solve structures at atomic resolution. This technique complements x-ray crystallography because it reveals structural details without the need for a crystalline specimen. Through the examination of a frozen-hydrated specimen in vitreous (non-crystalline) ice, the specimen ultrastructure, buffer composition, and ligand distribution from its native state are maintained. Cryo-EM also complements structural studies using nuclear magnetic resonance (NMR) in that it enables the study of specimens larger than 90 kDa. Structural biologists frequently use cryo-EM to study viruses, small organelles, and macromolecular biological complexes, purified proteins as well as molecular interactions in supramolecular assemblies or machines.
Single-particle cryo-electron microscopy (cryo-EM), is an increasingly popular technique used by structural biologists to solve structures at atomic resolution. This technique complements x-ray crystallography because it reveals structural details without the need for a crystalline specimen. Through the examination of a frozen-hydrated specimen in vitreous (non-crystalline) ice, the specimen ultrastructure, buffer composition, and ligand distribution from its native state are maintained. Cryo-EM also complements structural studies using nuclear magnetic resonance (NMR) in that it enables the study of specimens larger than 90 kDa. Structural biologists frequently use cryo-EM to study viruses, small organelles, and macromolecular biological complexes, purified proteins as well as molecular interactions in supramolecular assemblies or machines.