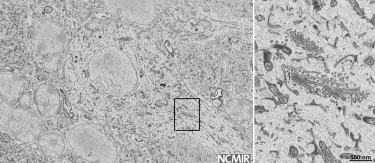
What is serial block-face imaging?
Serial block-face scanning electron microscopy (SBEM, SBSEM, and SBFSEM) is a way to reproducibly obtain high resolution 3D images from a sample. This method is particularly good at imaging large fields-of-view in X,Y,Z at nanometer resolution. SBEM typically relies on an in-situ ultramicrotome to be installed inside a scanning electron microscope (SEM). The SEM will collect an image of the block face, then the ultramicrotome will cut the sample to expose the next layer to be imaged, in steps as small as 15 nm.
Advantages of SBEM
| Capability | Advantage |
|---|---|
| Examines structure across orders of magnitude in scale | Maintains level of detail throughout a large dataset |
| Allows high throughput with no intervention | Useful to speed up sample analysis and reduce human error |
| Elucidates the context of observed ultrastructure | Enables you to gain more comprehensive results from one sample |
| Controls all aspects of the sectioning and acquisition process | Allows complete flexibility to optimize for a given sample |
| Eliminates section loss, damage and distortion | Avoids prohibitive processing to correct damage and distortion |
Uses
| Neuroscience | Cell biology | Material science | Other |
|---|---|---|---|
|
|
|
|
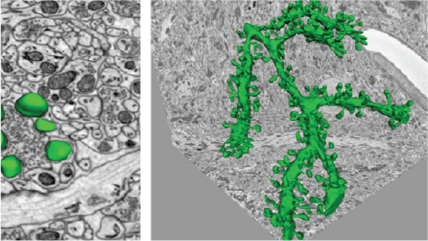
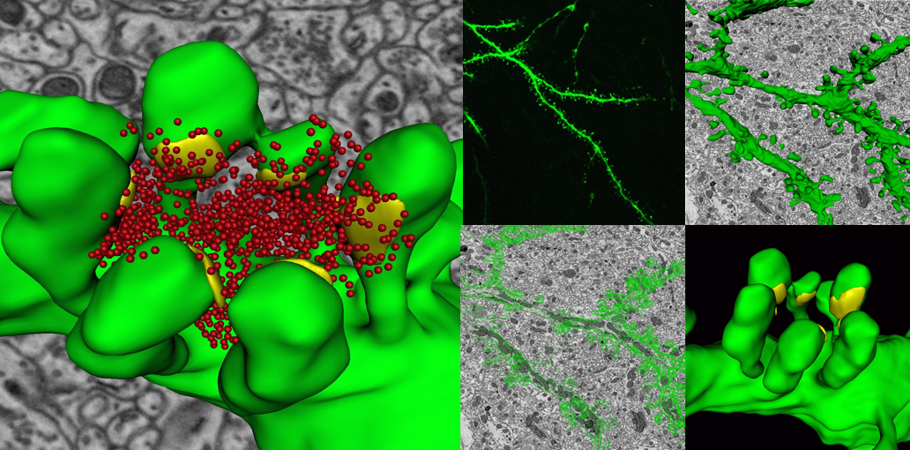
Neuroscience
A 3D reconstruction of a dendrite from a 15,625 μm³ (25 x 25 x 25 μm) volumetric data set containing 500 serial images of mouse cerebellum generated by the 3View® system. Dendrite structure (green), buttons (yellow), and vesicles (red). Inset images, clockwise from top left: Confocal image of a dendrite; wire frame traces rendered into a volumetric model; ultra-resolution dendritic spine model with synapses; and image showing wire frame traces.
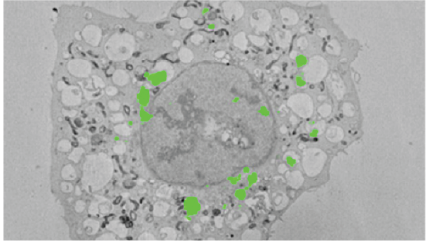
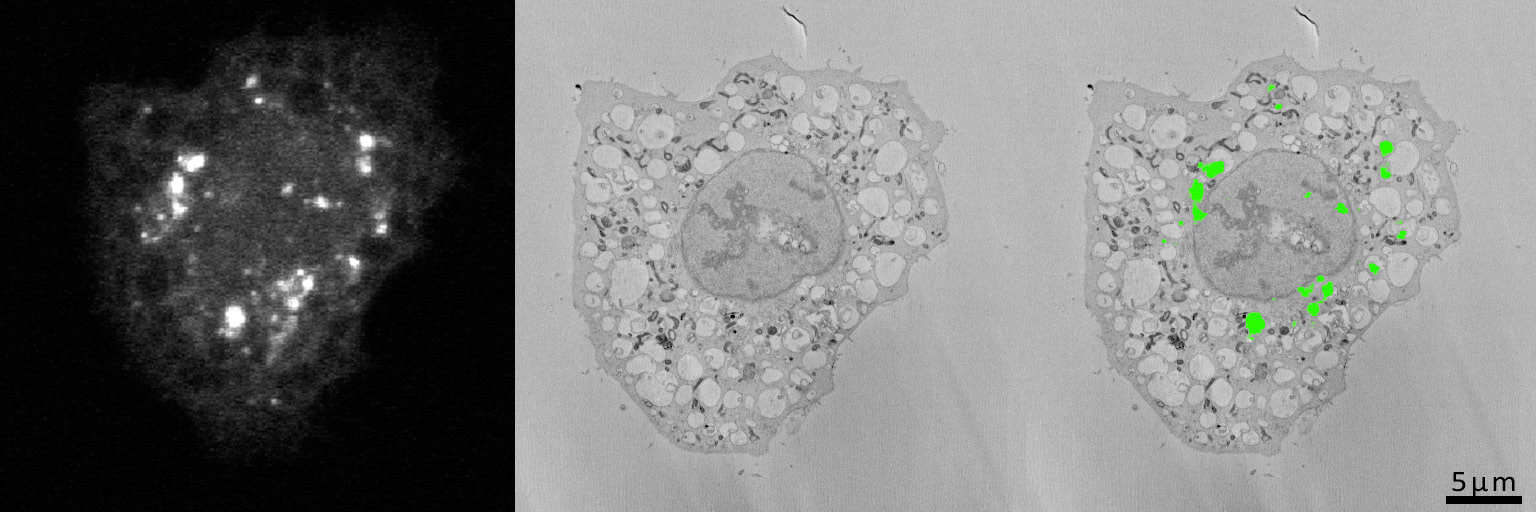
Cell biology: Complement confocal
Image of HeLa cells, stably expressing LC-GFP grown on gridded glass bottom coverslips dishes, starved for 2 h in serum-free medium and cells of interest identified by confocal microscopy. The cells were then processed in situ for electron microscopy and the coverslips dissolved from the epoxy resin with hydrofluoric acid. The cells were again identified in the resin block and in subsequent serial images generated by the 3View system.
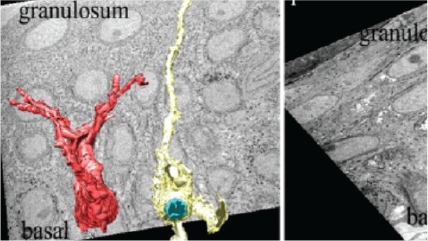
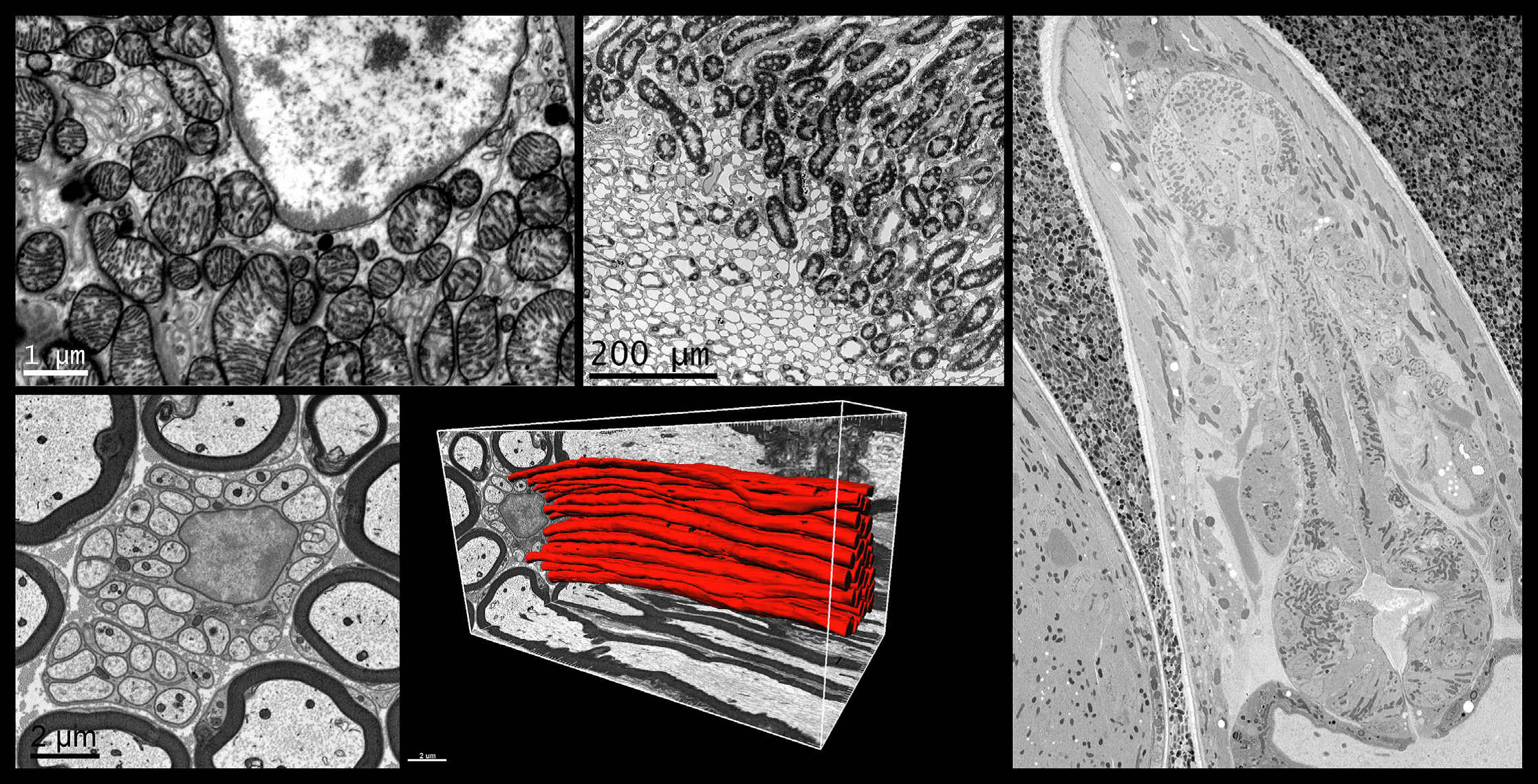
Cell biology: Developmental
Top left: High resolution, mouse kidney, 8192 x 8192 pixel image acquired with 1.5 nm pixels. Top middle: Large field-of-view, mouse kidney, 8192 x 8192 pixel image acquired with 80 nm pixels. Right: C. elegans prepared by high pressure freezing; 4096 x 4096 pixel image acquired with 25 nm pixels. Sample courtesy of Kent McDonald, University of California, Berkeley. Bottom left: Mouse sciatic nerve, 2048 x 2048 pixel image acquired with 5 nm pixels. Bottom middle: 3D visualization of mouse sciatic nerve axons.
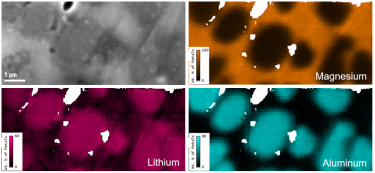
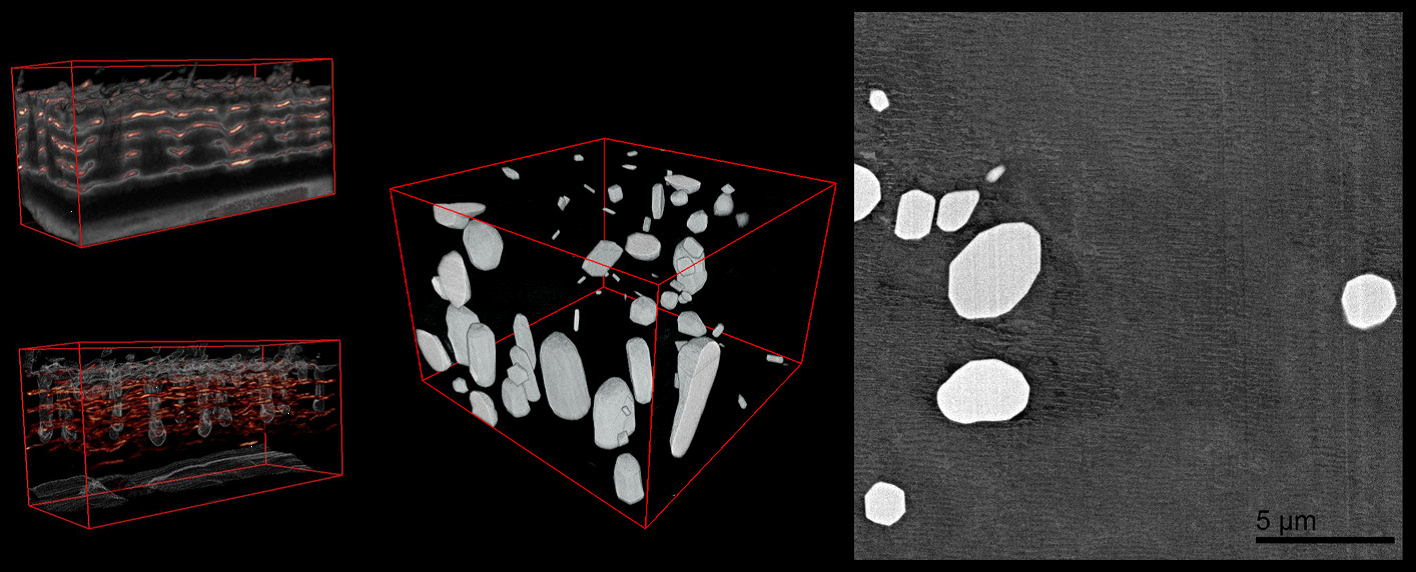
Material science
Upper and lower left: Images of anodized coating on aluminum surface generated by 3View system. Middle: 3D visualization of aluminum alloy with manganese particles generated by 3View system. 3D dataset contains one thousand 1024 x 1024 serial images with a pixel size of 15 nm and a cut thickness of 15 nm. 3D model created in DigitalMicrograph® using the 3D Visualization plugin.
Workflow for serial block-face imaging
3View system quick start guide
|
|
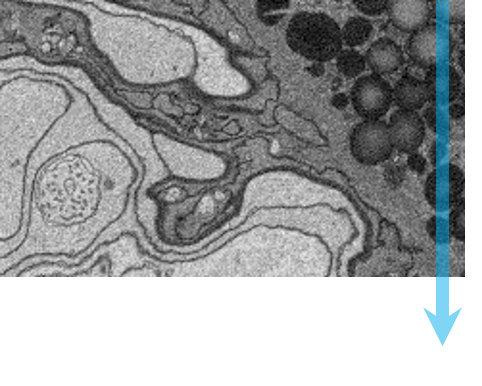
Step 1: Specimen preparation A typical biological specimen is fixed, stained with a contrasting agent and embedded in resin for stability. |
 |
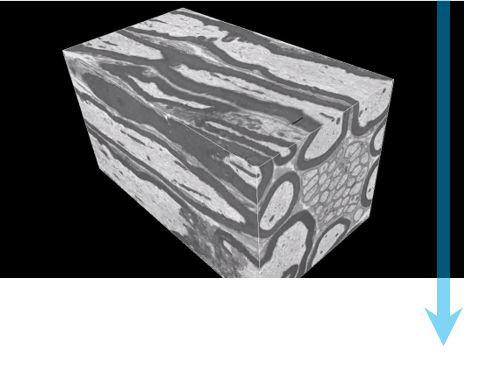
Step 2: Mount and transfer to SEM The specimen is trimmed to size, mounted to an aluminum pin, and optionally given a thin gold sputter coating. The specimen pin is put in the 3View system, the diamond knife is brought into contact, the door closed and evacuated. |
 |
Step 3: Optimize imaging The user selects suitable beam conditions, similar to conditions for standard SEM, but also considering effects in the Z direction. Magnification, pixel count, and dwell time are chosen such that the image has the desired resolution, field-of-view, and acquisition time. |
 |
Step 4: Automated imaging Sequential images are acquired with beam conditions of the user’s choice. Before each image, the knife removes a layer from the surface of the specimen. |
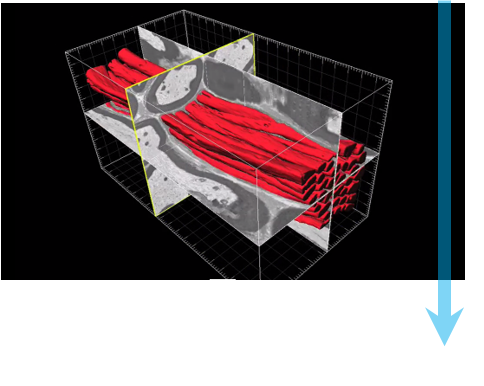 |
Step 5: Analyze The images are stacked to form a 3D dataset, which can be processed and viewed in DigitalMicrograph® or third-party software. At this point segmentation and quantification can be performed. |
Applications
|
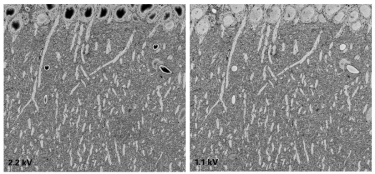
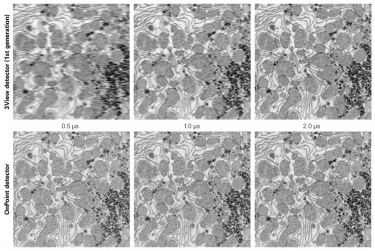
Optimizing three-dimensional electron microscopy results at low kV |
Protocols
3View recipes
3View quick start guide
3View diamond knife cleaning procedure
NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block-face scanning electron microscopy
OTO fixation for SEM and block-face imaging
Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization (Nature video)